Broad IBD Challenge
Back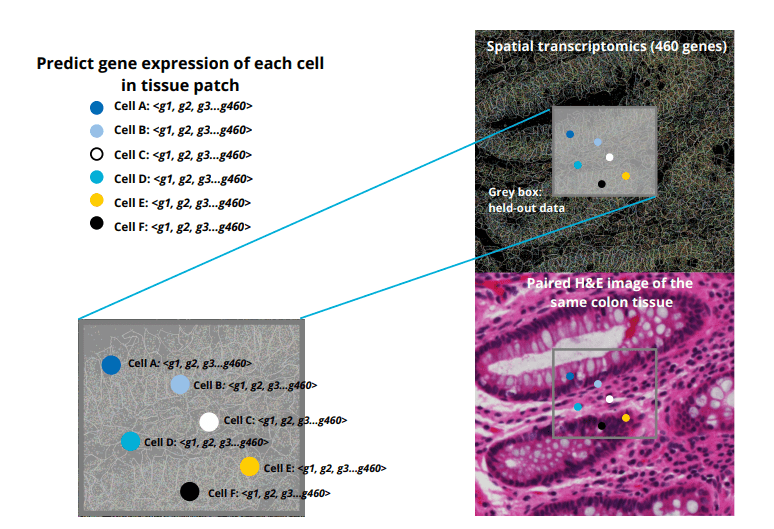
In the Autoimmune Disease Machine Learning Challenge organized by the Eric and Wendy Schmidt Center at the Broad Institute and partners, our team focused on Crunch 1, where the task was to predict spatial transcriptomics profiles from routine H&E pathology images. Specifically, we aimed to “inpaint” held-out patches of Xenium spatial transcriptomics data by leveraging the surrounding tissue’s matched H&E images and existing spatial transcriptomic measurements. This involved first aligning the H&E and DAPI-stained images via nuclear segmentation masks, then training a convolutional neural network—augmented with attention modules to capture both local cellular morphology and broader tissue context—to infer the expression of 460 genes in each nucleus of the held-out regions . Working alongside two partners, we preprocessed the provided SpatialData objects (including H&E registered images, segmentation masks, and per-cell gene counts) and implemented custom loss functions to balance global and local patch predictions. After two validation checkpoints, our model demonstrated robust inpainting capabilities across diverse colon tissue samples, from inflamed ulcerative colitis sections to reference diverticulitis tissue. Ultimately, our solution ranked 30th out of all participants in Crunch 1—an achievement that highlights both the complexity of high-resolution gene expression prediction and the promise of machine learning to bridge standard histology with spatial genomics.